rabvRedcapProcessing
Tools for processing rabies metadata for REDCap upload
Table of Contents
Introduction
rabvRedcapProcessing is an R package designed to streamline the cleaning, validation, and formatting of rabies lab metadata before uploading to REDCap. It automates common curation tasks including:
- Matching data to a REDCap dictionary
- Handling common mismatches and blank fields
- Recoding labels to dictionary codes
- Creating REDCap-compatible repeat instruments (diagnostic and sequencing forms)
This ensures data consistency across multiple labs and minimizes manual formatting errors.
Workflow
The typical workflow is illustrated below:
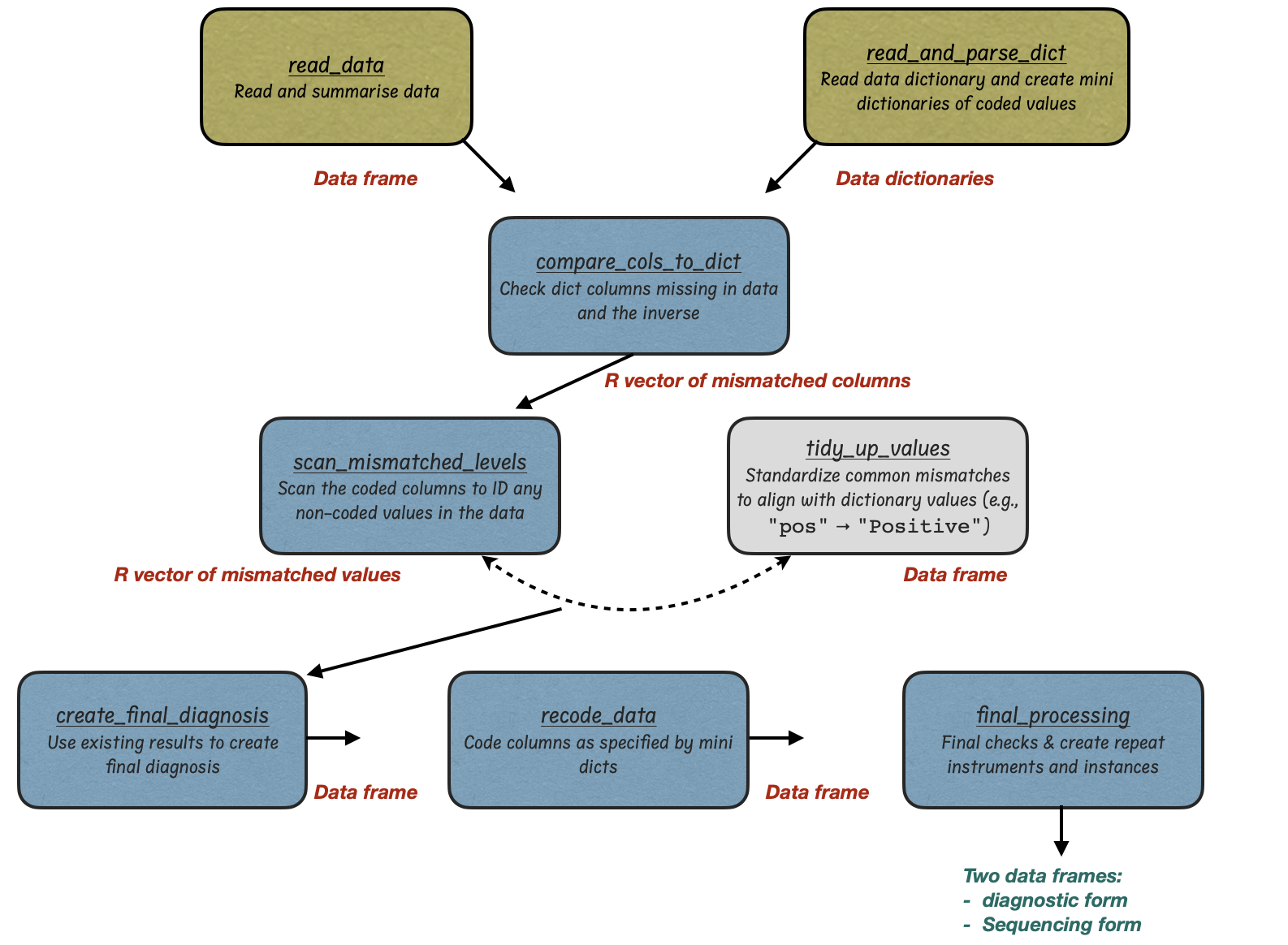
| Function | Description | Input | Output |
|---|---|---|---|
read_data() |
Load and summarize input metadata | csv filePath | Data frame + duplicates report |
read_and_parse_dict() |
Load REDCap dictionary and extract coded lists | optional dictPath | List of mini dictionaries |
compare_cols_to_dict() |
Check for missing or extra columns vs REDCap dictionary | Data + optionaldictPath | Harmonized data frame |
scan_mismatched_levels() |
Scan for values that don’t match allowed dictionary codes | Data + col_to_check + dicts | Warnings to inform your manual processing (if necessary) |
tidy_up_values() |
Fix common value mismatches (e.g., typos, synonyms) | Data | Cleaned data |
create_diagnostic_result_rule() |
Derive final diagnostic result from test columns | Data | updated diagnostic_result(df) |
recode_data() |
Recode labels to dictionary codes | Data + dicts | Recoded data frame |
final_processing() |
Final formatting & generation of REDCap-ready forms | Data + optionaldictPath + access_group | Diagnostic + Sequencing forms |
Installation
Install directly from GitHub using devtools:
# install.packages("devtools") # if not already installed
devtools::install_github("RAGE-toolkit/rabvRedcapProcessing")
library(rabvRedcapProcessing)
Bug reports
Please report issues or suggest improvements via GitHub:
URL: https://github.com/RAGE-toolkit/rabvRedcapProcessing
BugReports: https://github.com/RAGE-toolkit/rabvRedcapProcessing/issues
Contributors
Martha M. Luka, Kirstyn Brunker
